publications
2023
-
 Democratizing Computational Pathology: Optimized Whole Slide Image Representations for The Cancer Genome AtlasTristan Lazard , Marvin Lerousseau , Sophie Gardrat , and 5 more authorsDec 2023
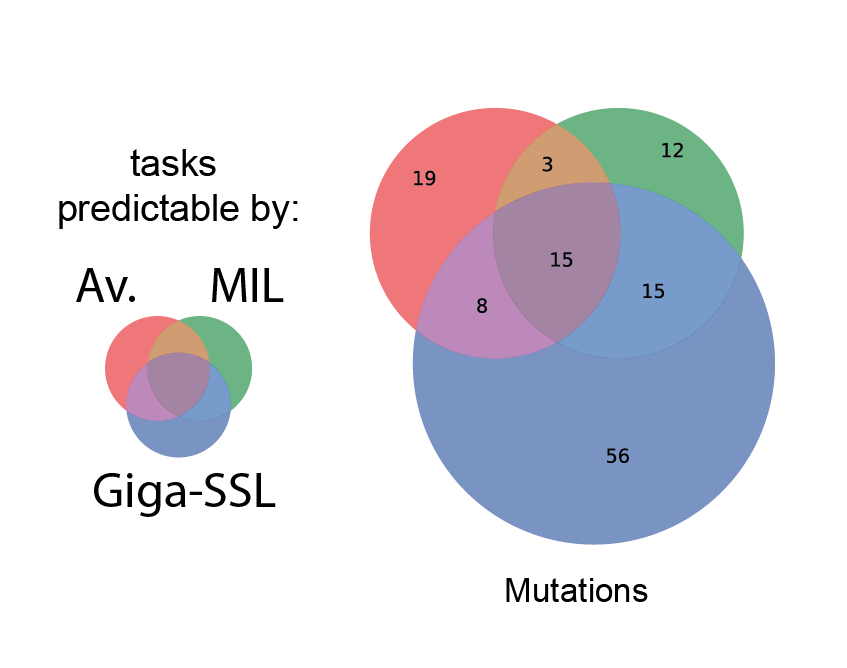
Democratizing Computational Pathology: Optimized Whole Slide Image Representations for The Cancer Genome AtlasTristan Lazard , Marvin Lerousseau , Sophie Gardrat , and 5 more authorsDec 2023Automatic analysis of hematoxylin and eosin (H&E) stained Whole Slide Images (WSI) bears great promise for computer assisted diagnosis and biomarker discovery. However, scarcity of annotated datasets leads to underperforming models. Furthermore, the size and complexity of the image data limit their integration into bioinformatic workflows and thus their adoption by the bioinformatics community. Here, we present Giga-SSL, a self-supervised method for learning WSI representations without any annotation. We show that applying a simple linear classifier on the Giga-SSL representations improves classification performance over the fully supervised alternative on five benchmarked tasks and across different datasets. Moreover, we observe a substantial performance increase for small datasets (average gain of 7 AUC point) and a doubling of the number of mutations predictable from WSIs in a pan-cancer setting (from 45 to 93). We make the WSI representations available, compressing the TCGA-FFPE images from 12TB to 23MB and enabling fast analysis on a laptop CPU. We hope this resource will facilitate multimodal data integration in order to analyze WSI in their genomic and transcriptomic context.
-
 Giga-SSL: Self-Supervised Learning for Gigapixel ImagesTristan Lazard , Marvin Lerousseau , Etienne Decencière , and 1 more authorIn 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW) , Jun 2023
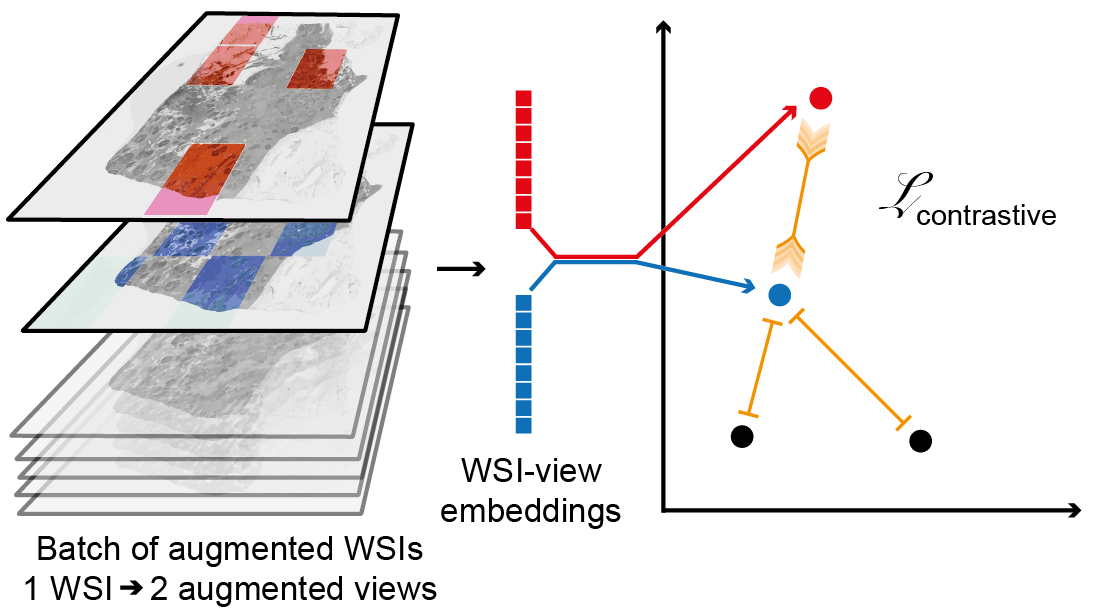
Giga-SSL: Self-Supervised Learning for Gigapixel ImagesTristan Lazard , Marvin Lerousseau , Etienne Decencière , and 1 more authorIn 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW) , Jun 2023 - Automatic Grading of Cervical Biopsies by Combining Full and Self-supervisionMélanie Lubrano , Tristan Lazard , Guillaume Balezo , and 4 more authorsIn Computer Vision – ECCV 2022 Workshops , Jun 2023
In computational pathology, predictive models from Whole Slide Images (WSI) mostly rely on Multiple Instance Learning (MIL), where the WSI are represented as a bag of tiles, each of which is encoded by a Neural Network (NN). Slide-level predictions are then achieved by building models on the agglomeration of these tile encodings. The tile encoding strategy thus plays a key role for such models. Current approaches include the use of encodings trained on unrelated data sources, full supervision or self-supervision. While self-supervised learning (SSL) exploits unlabeled data, it often requires large computational resources to train. On the other end of the spectrum, fully-supervised methods make use of valuable prior knowledge about the data but involve a costly amount of expert time. This paper proposes a framework to reconcile SSL and full supervision, showing that a combination of both provides efficient encodings, both in terms of performance and in terms of biological interpretability. On a recently organized challenge on grading Cervical Biopsies, we show that our mixed supervision scheme reaches high performance (weighted accuracy (WA): 0.945), outperforming both SSL (WA: 0.927) and transfer learning from ImageNet (WA: 0.877). We further shed light upon the internal representations that trigger classification results, providing a method to reveal relevant phenotypic patterns for grading cervical biopsies. We expect that the combination of full and self-supervision is an interesting strategy for many tasks in computational pathology and will be widely adopted by the field.
2022
-
 Deep Learning Identifies Morphological Patterns of Homologous Recombination Deficiency in Luminal Breast Cancers from Whole Slide ImagesTristan Lazard , Guillaume Bataillon , Peter Naylor , and 7 more authorsCell Reports Medicine, Dec 2022
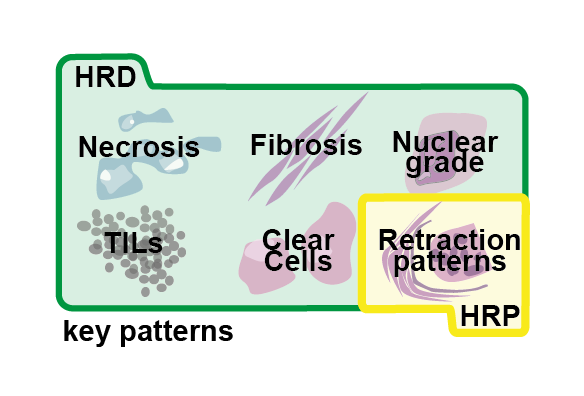
Deep Learning Identifies Morphological Patterns of Homologous Recombination Deficiency in Luminal Breast Cancers from Whole Slide ImagesTristan Lazard , Guillaume Bataillon , Peter Naylor , and 7 more authorsCell Reports Medicine, Dec 2022Homologous recombination DNA-repair deficiency (HRD) is becoming a well-recognized marker of platinum salt and polyADP-ribose polymerase inhibitor chemotherapies in ovarian and breast cancers. While large-scale screening for HRD using genomic markers is logistically and economically challenging, stained tissue slides are routinely acquired in clinical practice. With the objectives of providing a robust deep-learning method for HRD prediction from tissue slides and identifying related morphological phenotypes, we first show that digital pathology workflows are sensitive to potential biases in the training set, then we propose a method to overcome the influence of these biases, and we develop an interpretation method capable of identifying complex phenotypes. Application to our carefully curated in-house dataset allows us to predict HRD with high accuracy (area under the receiver-operator characteristics curve 0.86) and to identify morphological phenotypes related to HRD. In particular, the presence of laminated fibrosis and clear tumor cells associated with HRD open new hypotheses regarding its phenotypic impact.
- Prediction of Treatment Response in Triple Negative Breast Cancer From Whole Slide ImagesPeter Naylor , Tristan Lazard , Guillaume Bataillon , and 5 more authorsFrontiers in Signal Processing, Dec 2022
The automatic analysis of stained histological sections is becoming increasingly popular. Deep Learning is today the method of choice for the computational analysis of such data, and has shown spectacular results for large datasets for a large variety of cancer types and prediction tasks. On the other hand, many scientific questions relate to small, highly specific cohorts. Such cohorts pose serious challenges for Deep Learning, typically trained on large datasets. In this article, we propose a modification of the standard nested cross-validation procedure for hyperparameter tuning and model selection, dedicated to the analysis of small cohorts. We also propose a new architecture for the particularly challenging question of treatment prediction, and apply this workflow to the prediction of response to neoadjuvant chemotherapy for Triple Negative Breast Cancer.